The week-long International Conference on Machine Learning (ICML) ended June 24, and the last day included the 2016 ICML Workshop on Computational Biology. CSE professors Larry Smarr and Rob Knight as well as Qualcomm Institute data scientist Mehrdad Yazdani were represented in a poster presentation and paper on "Using Topological Data Analysis to find discrimination between microbial states in human microbiome data." Borrowing a statistical method originally from topology, the co-authors applied Topological Data Analysis (TDA) as an "unsupervised learning and data exploration tool to identify changes in microbial states."
"Since the human microbiome ecology differs dramatically in different body sites [parts] and individuals," said Yazdani, "understanding how and what changes in the ecology are of crucial importance."
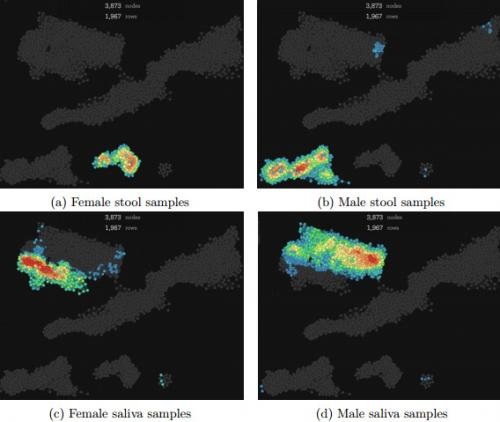
 Yazdani works closely with Smarr and Knight -- whose appointments are in Pediatrics and CSE -- on analyzing colonies of species in the human microbiome in healthy and sick subjects, notably for Smarr's Future Patient project. To test the TDA method, they used a previously published dataset of high-resolution time series of the microbiome from three different sites (mouth, hands, and gut) and from two healthy subjects (one female, one male). Previous studies have shown that microbial communities of a healthy subject are highly stable over time, so TDA and other methods should have been able to identify six total microbial communities - three for the male subject based on his different body sites, and three for the female subject's three sites.
Yazdani works closely with Smarr and Knight -- whose appointments are in Pediatrics and CSE -- on analyzing colonies of species in the human microbiome in healthy and sick subjects, notably for Smarr's Future Patient project. To test the TDA method, they used a previously published dataset of high-resolution time series of the microbiome from three different sites (mouth, hands, and gut) and from two healthy subjects (one female, one male). Previous studies have shown that microbial communities of a healthy subject are highly stable over time, so TDA and other methods should have been able to identify six total microbial communities - three for the male subject based on his different body sites, and three for the female subject's three sites.
The scientists wanted to see how TDA compared to other well-established methods, PCA and MDS*. The older methods did identify the clusters for three sites, but did not detect a difference based on the subject's gender. "These methods [PCA and MDS] do not discriminate samples based on the subjects," they noted in the paper. On the other hand, the TDA method identified distinct clusters that discriminated between the female and male gut samples, and based on the skin and tongue body sites. Concluded Yazdani: "This suggests that TDA is able to identify groups of clusters that other methods may potentially miss."
The ICML Workshop on Computational Biology brought together researchers applying machine learning to challenging biological questions, especially given the development of high-throughput technologies such as next-generation sequencing, mass cytometry (CyTOF), and single-cell sequencing, all of which can now generate vast amounts of data from the biological systems in question.
___________________________________
*PCA stands for Principal Component Analysis; MDS refers to Multi-Dimensional Scaling (also known as Principal Coordinate Analysis).
Read the Using Topological Data Analysis paper by Yazdani, Smarr and Knight.
ICML 2016 Workshop on Computational Biology

