
UC San Diego hosts mini-symposium to coincide with project report in the journal Nature

Rob Knight addresses the Earth Microbiome
Project Mini-Symposium.
The Center for Microbiome Innovation (CMI), led by Pediatrics and Computer Science and Engineering professor Rob Knight, hosted a mini-symposium Nov. 1 in the Qualcomm Institute. Knight welcomed attendees at the first public presentation of preliminary findings of the Earth Microbiome Project, and the mini-symposium was timed to coincide with the publication in the journal Nature of results from work undertaken by the international effort begun in 2010. The paper was co-authored by more than 300 researchers at over 160 institutions worldwide.
The Earth Microbiome Project (EMP) is a large-scale study of microbial diversity based on the analysis of DNA sequences from samples submitted by scientists from around the globe. The project is building an open-access resource that Nature dubbed an “experimental tour de force.” EMP is co-led by researchers at the University of California San Diego, Pacific Northwest National Laboratory, University of Chicago, and Argonne National Laboratory. (UC San Diego also houses the American Gut Project, a crowdsourcing effort that grew out of the EMP, according to CMI and American Gut director Knight.)

Earth Microbiome Project published in Nature.
The deans of both the Jacobs School of Engineering and the UC San Diego School of Medicine gave opening remarks, but the task of presenting an overview of the Earth Microbiome Project fell to the CMI director. UC San Diego’s Knight noted that “Earth is a microbial planet, with 100 million times as many bacteria on Earth than there are stars in the universe. That makes microbiomes the largest data set in any science.” Microbiomes are unique collections of microbes living in each sample.
“Microbes are everywhere,” said first author Luke Thompson, a former researcher in Knight’s lab who is now a research associate at the National Oceanic and Atmospheric Administration (NOAA). “Yet prior to this massive undertaking, changes in microbial community composition were identified mainly by focusing on one sample type, one region at a time. This made it difficult to identify patterns across environments and geography to infer generalized principles.”
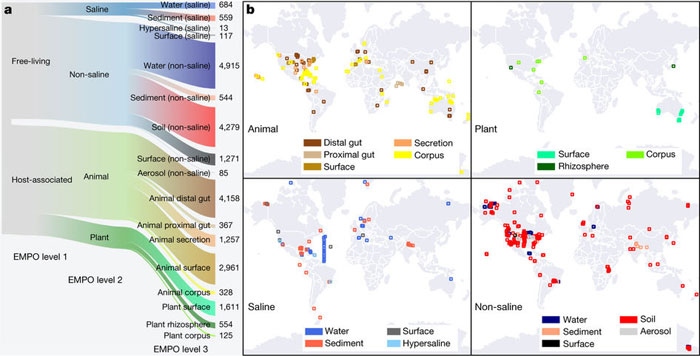
Knight confirmed that the preliminary results published in Nature Nov. 1 are based on more than 27,000 samples collected via crowdsourcing from a vast number of different environments from 43 countries around the globe. (The samples came from 97 independent studies involving a wide range of geographic and environmental ecosystems, both on land and in marine environments.)
According to Knight, the scientific community really got behind the project. “Prior to this effort, the scientific literature only documented 202 environmental samples, for a total of 21,000 microbial DNA sequences. Now, we’ve collected more than 27,000 samples for a total of 2.2 billion sequences,” he said. “This highlights one of the benefits of open-source scientific data. We wouldn’t have been able to acquire these data any other way.”
Project researchers generated the 2.2 billion DNA sequences for a highly variable region of a gene called 16S rRNA (a gene that encodes a component of the cell’s protein synthesis machinery, the ribosome). The combined sequences represent the first reference database of bacteria covering the planet, and the paper represents the first major milestone toward the Earth Microbiome Project’s long-term vision of constructing “the microbial map for planet Earth.”
“The remarkable nature of this study lies not only in its scale and in the breadth of the environmental samples analyzed, but also in its methodology,” noted Nature writer Jeroen Raes in an article released in connection with the new paper. “The project involved a massive, global crowdsourcing effort in which scientists raided their collection freezers for samples to share with the project.”
After analyzing these microbiomes, the project generated 2.2 billion DNA sequence ‘reads’ of a highly variable region of a gene called 16S rRNA (a gene that encodes a component of the cell’s protein synthesis machinery, the ribosome). The combined sequences represent the first reference database of bacteria covering the planet, and publication of the findings in Nature represents the first major milestone toward EMP’s long-term vision of constructing a “microbial map for planet Earth.”
According to Knight, the project is now poised to continue to grow and generate new scientific discoveries as new data are added, and scientists conform to newly standardized protocols, original analytical methods and open data-sharing. In his opening talk, Knight noted that although the preliminary study involved 27,000 samples, the Earth Microbiome Project has actually collected some 100,000 samples to date.
"There are large swathes of microbial diversity left to catalogue, and yet we’ve ‘recaptured’ about half of all known bacterial sequences,” said Jack Gilbert, who directs the Microbiome Center at the University of Chicago, who followed Knight at the mini-symposium podium. “With this information, patterns in the distribution of the Earth’s microbes are already emerging.”

Mini-Symposium on November 1.
Another speaker – Janet Jansson, chief scientist at the Pacific Northwest National Laboratory – noted that “these global ecological patterns offer just a glimpse of what is possible with coordinated and cumulative sampling. More sampling is needed to account for factors such as latitude and elevation, and to track environmental changes over time. The Earth Microbiome Project provides both a resource for the exploration of myriad questions, and a starting point for the guided acquisition of new data to answer them.”
Other speakers included University of Colorado, Boulder professor Val McKenzie, and four researchers who gave lightning talks about some of the roughly 100 research projects already carried out as part of the Earth Microbiome Project. Data made available through the EMP has already been used, for example in studies of drinking water, oil spills, the gut health of primates, how soil microbes affect wine grapes, how obesity affects smelling of wine aromas, and how the human microbiome changes over time and alters the spaces where we live.
The two-hour symposium was webcast in real time from the Qualcomm Institute and distributed via Facebook Live. An archive record of the two-hour event will shortly be available for on-demand viewing on the Facebook page of the Center for Microbiome Innovation.
____________________________________
*Luke R. Thompson… Rob Knight & Earth Microbiome Project Consortium, A Communal Catalogue Reveals Earth’s Multiscale Microbial Diversity, Nature, 1 November 2017. (doi:10.1038/nature24621)
Related Links
Earth Microbiome Project
Mini-Symposium Archived Webcast from Facebook Live
Full Nature Paper
Center for Microbiome Innovation

